dbAMEPNI: A database of Alanine Mutagenic Effects for Protein-Nucleic Acid Interactions
dbAMEPNI provides a user friendly interfaces to facilitate the viewing of the data, this website allows for browsing the datasets and provides a quick search for flexible query. dbAMEPNI contains 859 mutant information, all of the information are deposited from 217 original research articles. Mutagenesis and binding data can be downloaded directly from the dbAMEPNI web site. In the future, dbAMEPNI will continue to be updated. To keep the database current, submission of new data is encouraged. Thanks for your devotion!
The dbAMEPNI requires a modern web browser with JavaScript and cookies enabled. To view the complex details, pop-ups must not be blocked.The following browsers have been throughly tested with dbAMEPNI:
- Mozilla Firefox, version 4 or above
- Chrome, version 5 or above
- Internet Explorer, versions 7, and 8 or above
The latest version of Firefox and Chrome is recommended for visualization.
PDBID: A 4-character unique identifier of a protein-nucleic acid complex deposited in the Protein Data Bank
CH: Chain IDs of the protein-nucleic acid complexes
NAType: The nucleic acid type in the protein-nucleic acid complex
Mutant: The information about the original residue type and the residue's sequence number
WT_Kd: The dissociation constant of the wild type protein and nucleic acids
Mut_Kd: The dissociation constant of the mutated protein and nucleic acids
ΔΔG: The binding affinity difference of the complex between mutated state and wild type state. Note we assigned a large value of 5 kcal/mol for those cases whose binding affinity for the mutated state is "no binding" by the corresponding experiment.
Method: The experimental technology used to measure thermodynamic data
Temp: The temperature under which the thermodynamic data were measured
Sources: Where the data was collected
PMID: The unique identifier of a reference in PUBMED database
Pages: The specific page or table or figure of the reference where the data was collected
ASAbnd: The solvent accessible surface area of the residue in bound state calculated by NACCESS
ASAubd: The solvent accessible surface area of the residue in unbound state calculated by NACCESS
Hbond: The number of hydrogen bonds between the wild type residues and the nucleic acids determined by WHAT IF
SS: The secondary structure that the residue belongs to, which was calculated by DSSP program (downloaded from http://swift.cmbi.ru.nl/gv/dssp/) from PDB entries.
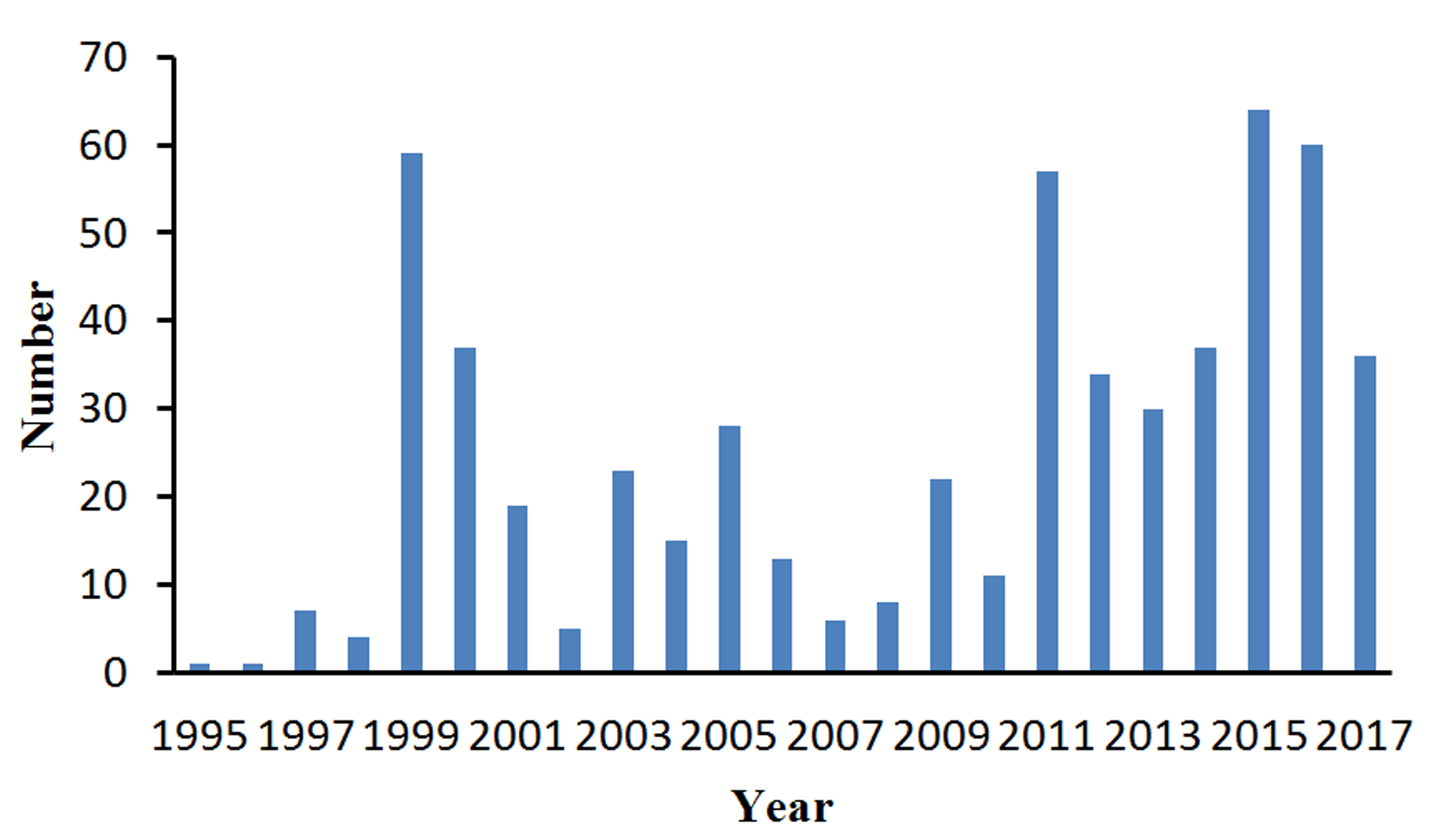
Yearly Growth of Residues in the Coreset

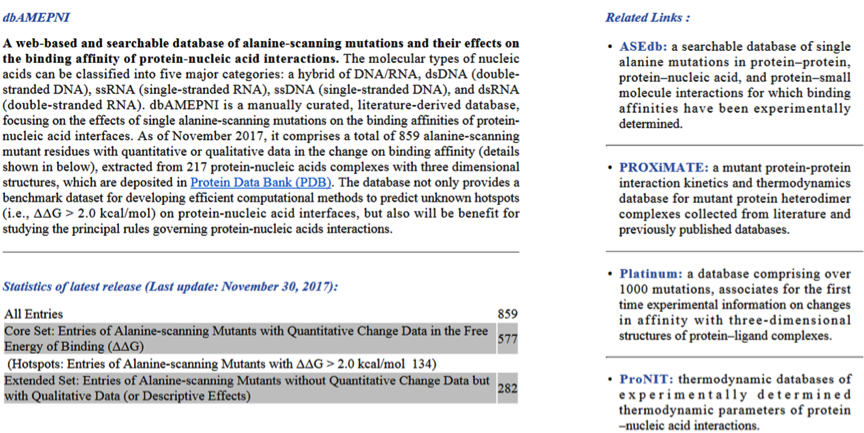
Please enter the URL http://210.45.212.102/dbAMEPNI/index.php in your browser to visit the homepage of dbAMEPNI, there are some brief introduction and a simple statistics of dbAMEPNI.

In the Browse page, you can see the two datasets, you can choose the one you need, and clicking the type, you will find more details information of this type.
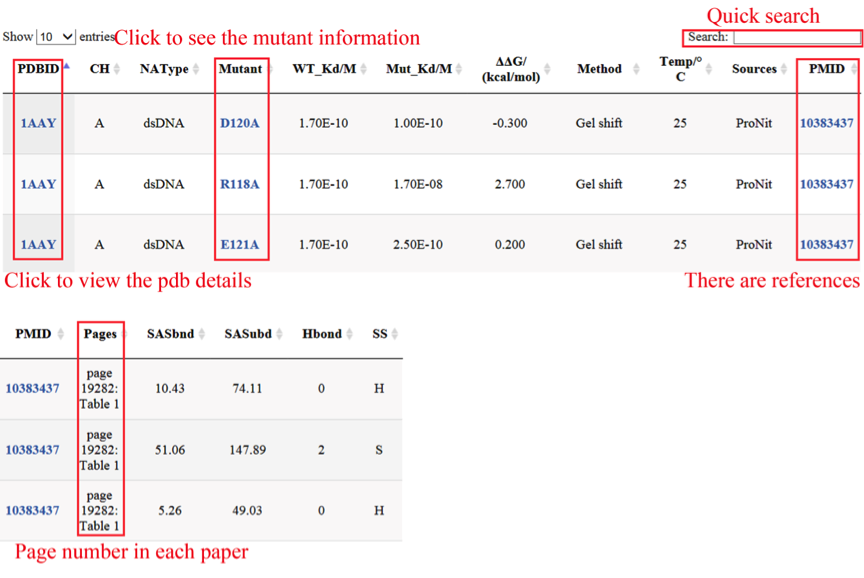
A full (or query) list of the benchmarking sets integrated within a panel is displayed in the details page. You will find a series of information, such as PDBID, mutant information, Thermodynamic data, PMID, solvent accessible surface area in unbound(SASubd) and bound(SASbnd) state, hydrogen bond(Hbond) between residue and nucleic acid and secondary structure (SS) and so on. And in this page ,you can use the quick search in the upper right corner of the page. you can provide a PDBID, and then will feedback a result of this PDBID in our database.


In the Search page, the users can query the database by specifing three main terms. First, the PDBID, the users can input a PDBID and get all entries related to the PDBID. Second, the maximum △△G and the minimum △△G, through this way, the users can obtain the entries of which the △△G are in certain range. Third, Method, the user query the entries for which the thermodynamic data were measured by a specific experimental method.


In this submit page, users can deposit a file or write a detail information about a mutation to us ,we will check it and edited for proper formation to added in dbAMEPNI. Thanks for your devotion.


In this download page, you will see the all three parts of protein-nucleic acid interactions data, you can freely download our data for academic researchers, but not for profit purposes.
If you have any suggestion to add new comment to records in current dbAMEPNI or to revise wrong information in current dbAMEPNI , please send us email directly.