m5CPred-SVM: A webserver for predicting m5C sites of RNA based on SVM
The m5CPred-SVM Server is a web-based implementation of m5CPred-SVM model – a machine learning approach for 5-methylcytosine (m5C) sites identification of RNA. The outputs of m5CPred-SVM are easy to understand, and could be browsed online or downloaded for further study. The results obtained by m5CPred-SVM on the three species: H. sapiens, M. musculus and A. thaliana indicate that the current predictor may become a useful high-throughput tool for predicting m5C sites.
The m5CPred-SVM model was developed by using support vector machine based on position specific propensity features such as PSNP and PSDP and nucleotide composition features such as KNF(K‑nucleotide frequencies), KSNPF(K-spaced nucleotide pair frequencies), and PseDNC(Pseudo dinucleotide composition).
The m5CPred-SVM server requires a modern web browser with JavaScript and cookies enabled. The following browsers have been throughly tested with m5CPred-SVM:
- Mozilla Firefox, version 4 or above
- Chrome, version 5 or above
- Internet Explorer, versions 7, and 8 or above
The latest version of Firefox and Chrome is recommended for visualization.

Analysis Procedure
To predict the possible m5C sites, following the steps below:.
• Select a species (red ‘2’ in the above figure) before you start to run a prediction.
• Submit your FASTA sequence (red ‘3’ in the above figure). You can also click the example button (red ‘1’ in the above figure) to get an example.
• Click button ‘Submit’ to submit request to m5CPred-SVM, or click ‘Clear’ button to clear all sequences (red ‘4’ in the above figure). The result will be shown in a new page.
• The submitted RNA sequence must be longer than 41 bp in FASTA format.
• If there is no cytosine residue in the submitted RNA sequence, the m5CPred-SVM server will have no output.
• The submitted RNA sequence cannot contain any illegal character. All but A, U, C and G are illegal characters
• If you make above mistake, you will get prompt message or warning message in the download file.
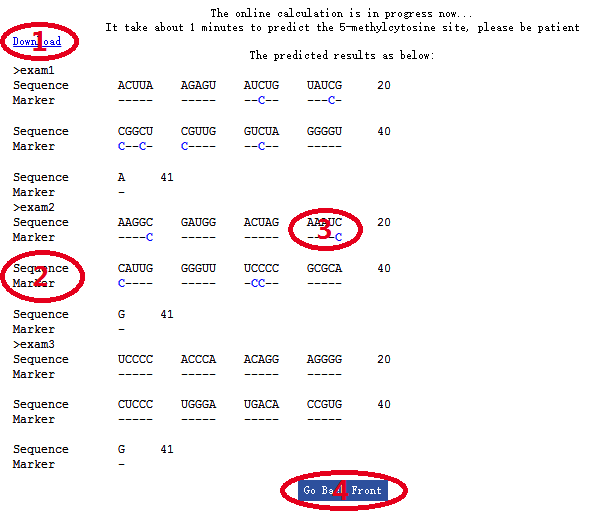
If you submit the sequence from the example, the output is shown as in the above figure. There are two rows to show the results: ‘Sequence’ and ‘Marker’ (red ‘2’ in the above figure). In the ‘sequence’ row, the original query sequence is displayed. In the ‘Marker’ row, the position that the nucleotide in the original query sequence that predicted for m5C site is highlighted in blue color (red ‘3’ in the above figure). If you want to continue to predict other sequence, please click ‘Go Back Front’ (red ‘4’ in the above figure) button to return to the homepage.

If you want to download the predicted results, the ‘download’ button can be found in red ‘1’ in the figure of Results section. There is more detailed information that predicted to be the m5C site. The above figure shows the predicted results for the sequence from the example, the first red box (red ‘1’ in the above figure) indicates the location of m5C sites in the whole query sequence, and the second red box (red ‘2’ in the above figure) represents the serial number of the query sequence.
If you have any suggestion or problem about m5CPred-SVM server , please send us email directly at xlzhu_mdl@hotmail.com.