iPPHOT: A webserver for predicting hotspots at protein-protein interfaces
The iPPHOT Server is a web-based implementation of iPPHOT model — a machine learning approach for predicting binding hot spots. The server facilitates the automated analysis of a given chain of protein and the visualization of its hot spot predictions. For each residue within the interface, the iPPHOT Server characterizes relative consevation, solvent accessible surface area and B factors of the residue, and predicts if the residue is a hot spot. The user can visualize the residue by clicking the corresponding row of the table after the computational analysis is complete.
The iPPHOT model is built by the combination of 3 algorithms: decision tree, mRMR and PSFS(Pseudo Sequential Forward Selection). For this work, hot spots are defined as mutations associated with a change in binding energy (∆∆G) greater than 2 kcal/mol.
The iPPHOT server requires a modern web browser with JavaScript and cookies enabled. To view the complex details, pop-ups must not be blocked.The following browsers have been throughly tested with iPPHOT:
- Mozilla Firefox, version 4 or above
- Chrome, version 5 or above
- Internet Explorer, versions 7, and 8 or above
The latest version of Firefox and Chrome is recommended for visualization.
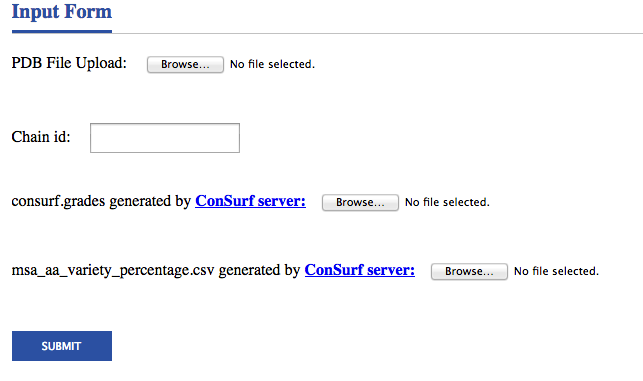
Before the iPPHOT analysis can begin, a user must provide a PDB file that contains the structure of a protein complex, a chain of the protein-protein interface for analysis, a consurf.grades file and a msa_aa_variety_percentage.csv file that are both generated by ConSurf server. Files that do not contain a bound complex are unlikely to yield useful results. In addition, model structures containing many clashes may vastly overestimate the number of hot spots. Finally, iPPHOT is able to analyze proteins but not other types of molecules. Please remove these from your PDB file before submitting to the server.
To analyze a chain of a protein-protein interface, enter the following information on the submission page and click the SUBMIT button.
• PDB File Upload — Link to PDB file of a protein complex (e.g. "1a4y.pdb")
• Chain id — A chain of a protein-protein interface for analysis (e.g. "A")
• consurf.grades generated by ConSurf server — File containing conservation scores generated by ConSurf server (e.g. "consurf1a4yA.grades")
• msa_aa_variety_percentage.csv generated by ConSurf server — File containing the residue variety in % for each position in the query sequence generated by ConSurf server (e.g. "msa_aa1a4yA_variety_percentage.csv")
Upon submission, the task will wait for processing. After processing begins, a typical iPPHOT analysis finishes within a minute. When the task is complete, a result is displayed to the user with their iPPHOT hot spot predictions or an error message. If the job finishes successfully, the status field will contain a table of results. Jobs that end in error are described by the following warnings:
• Your PDB file could not be uploaded because: No file was uploaded.
• Please assign a chain ID of your PDF file.
• Your consurf.grade file could not be uploaded because: No file was uploaded.
• Your msa_aa_variety_percentage.csv file could not be uploaded because: No file was uploaded.
Users can access an iPPHOT output by clicking the “Download” button. Most errors are caused by non-standard amino acids or ligands incorrectly labeled as ATOM records within the PDB coordinate file. If possible, the user should resolve the inconsistencies in the file and submit a new job. If subsequent jobs still end in error, users can contact xlzhu_mdl@hotmail.com for assistance.
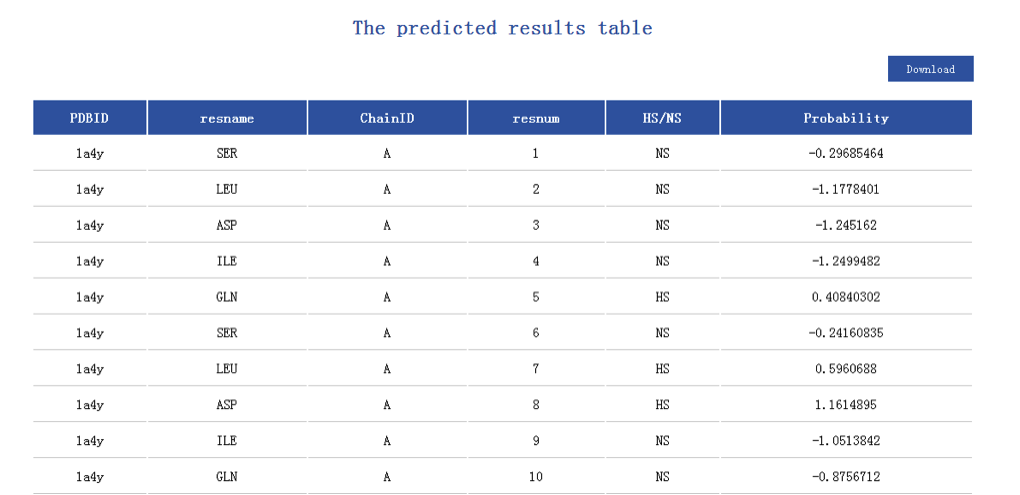
• PDBID — Protein identifier from PDB file.
• resname — Amino acid residue name.
• ChainID — Chain identifier chosen from PDB file.
• resnum — Residue number from PDB file.
• HS/NS — Hot spot(HS)/non-hot spot(NS) predicted as by iPPHOT.
• Probability — Probability of prediction by iPPHOT.
• Download — On the uperright, users can click download to downlaod all the results.
Users can directly click the corresponding row to view a residue in the table. Meanwhile, scaling and spin are available for further observation.
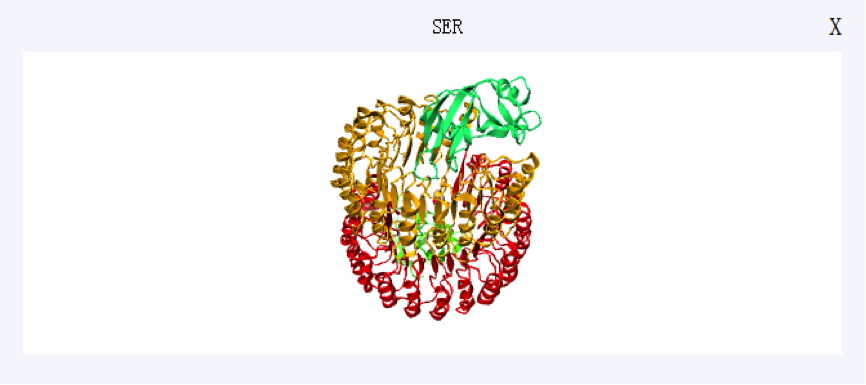
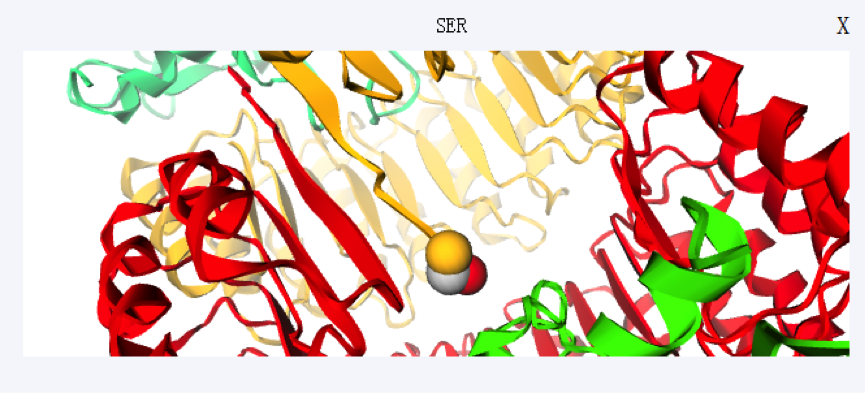

If you have any suggestion or problem about iPPHOT server , please send us email directly.