ATMGBs: A webserver for predicting protein binding sites of protein-nucleic acid complexes
The ATMGBs Server is a web-based implementation of ATMGBs model — is a novel deep learning framework for sequence-based prediction of protein-nucleic acid binding sites. It integrates multiple feature types and leverages attention maps from protein language models to simulate spatial relationships between residues. It needs only one input: a sequence in fasta format. Note, due to the limited resources of the server, it can only deal with one sequence each time.
The ATMGBs model is built by integrate the embedding of two protein language models, ESM1B and ProtT5.
The ATMGBs server requires a modern web browser with JavaScript and cookies enabled. To view the complex details, pop-ups must not be blocked.The following browsers have been throughly tested with ATMGBs:
- Mozilla Firefox, version 4 or above
- Chrome, version 5 or above
- Internet Explorer, versions 7, and 8 or above
The latest version of Firefox and Chrome is recommended for visualization.
Before the ATMGBs analysis can begin, a user must provide a fasta file that contains the sequence of only one protein, and specify which kind of nucleic acid it may bind.
To analyze binding sites of a protein, enter the following information on the submission page and click the SUBMIT button.
• Sequence in Fasta format
Upon submission, the task will wait for processing. After processing begins, a typical ATMGBs analysis finishes within a minute. When the task is complete, a result is displayed to the user with their ATMGBs binding sites predictions or an error message. If the job finishes successfully, the status field will contain a table of results. Jobs that end in error are described by the following warnings:
• Your sequences cannot be blank.
Users can access an ATMGBs output by clicking the “Download” button. Most errors are caused by non-standard amino acids or ligands incorrectly labeled as ATOM records within the PDB coordinate file. If possible, the user should resolve the inconsistencies in the file and submit a new job. If subsequent jobs still end in error, users can contact xlzhu_mdl@hotmail.com for assistance.
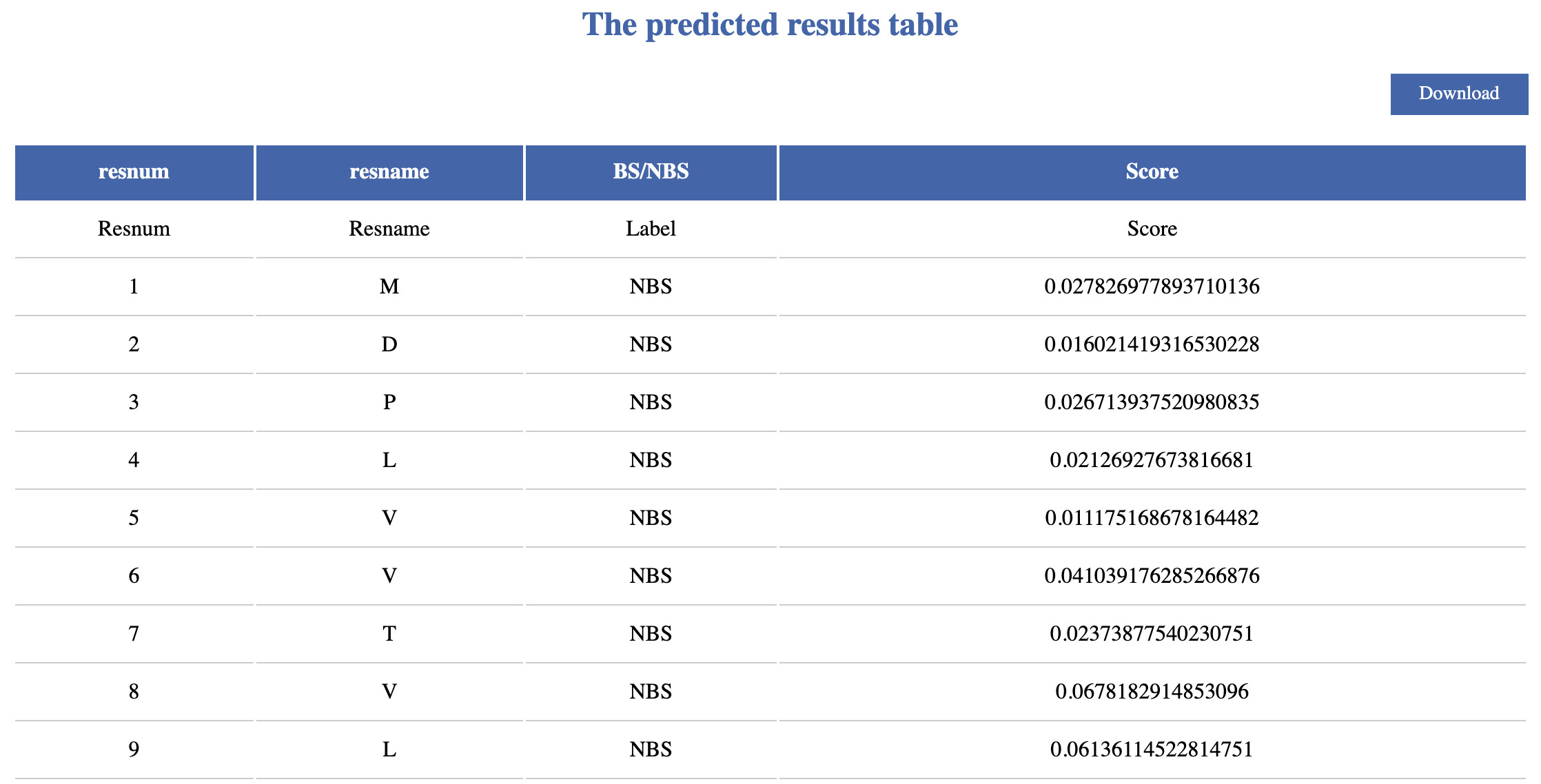
• resnum — Residue number of the protein.
• resname — Amino acid residue name.
• BS/NBS — Binding Sites(HS)/non-Binding Sites(NS) predicted as by ATMGBs.
• Scores — Probability of prediction by ATMGBs.
• Download — On the uperright, users can click download to downlaod all the results.

If you have any suggestion or problem about ATMGBs server, please send us email directly.